Our Science
XenoAptamers
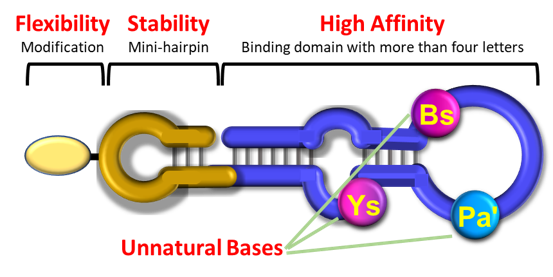
Xenolis’ new XenoAptamers, short single-stranded DNA or RNA with our proprietary unnatural bases, enhance binding to target molecules like proteins and cells with increased affinity and specificity. The addition of a mini-hairpin sequence stabilizes XenoAptamers against heat and nucleases, also serving as a modification site.
XenoAptamers are generated by the ExSELEX platform, an innovative evolutionary engineering technique that utilizes a six-letter DNA alphabet. Once the optimal XenoAptamer sequence is identified, it can be chemically synthesized for large-scale production and site-specific modifications. This surpasses traditional antibodies by providing superior quality control, exceptional affinity and specificity, low immunogenicity, and improved water solubility.
References
Dengue diagnostics:
ELISA:
Our Advantages
Improved Binding Affinity
Hydrophobic unnatural bases boost aptamer affinity, enhancing sensitive and selective protein detection.
Adjustable Specificity
XenoAptamers can be engineered to recognize amino acid sequence variations or multiple mutant proteins.
High Bio-Stability
Stable across diverse conditions, XenoAptamers ensure consistent results, protected from degradation by a proprietary mini-hairpin DNA.
Site-Specific Modification
Easily modified for targeted applications, the mini-hairpin region is ideal for attaching functional groups like fluorescent dyes, biotin, or drugs. a
Ability-Specific Binding
XenoAptamers selectively bind to specific protein regions or isoforms, even those usually inaccessible to antibodies.
Effective on ‘Hard-to-Target’ Glycosylated Proteins
XenoAptamers retain high affinity to glycosylated protein target.
Low Toxicity and Immunogenicity
With minimal unnatural bases, XenoAptamers exhibit low toxicity and immunogenicity.
Cost-effective
They are inexpensive and scalable, making them an economical choice for large-scale use.
Effective XenoAptamer-Antibody Sandwich Pair
ExSELEX with a capture antibody produces XenoAptamers that form the most effective and optimal XenoAptamer-antibody sandwich pairs for detecting targets and biomarkers using ELISA and LFA.
The ExSELEX™ Platform
TM
selection and development services for a wide range of life science applications.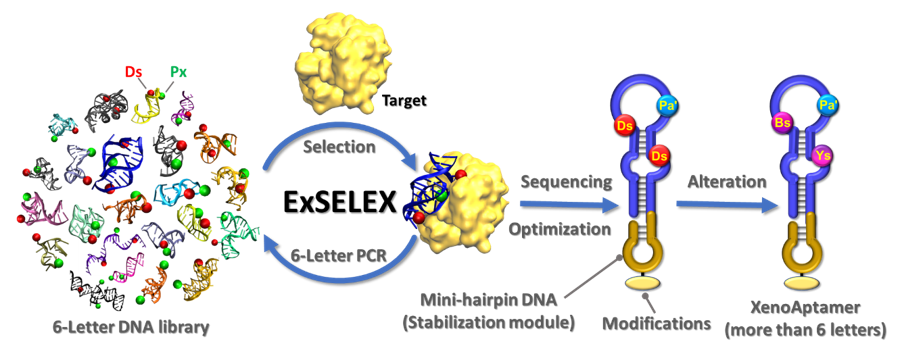
References
Success Rates Of XenoAptamer Generation:
Alteration process:
Initial Technology:
Mini-hairpin XenoAptamer:
Mini-Hairpin DNA:
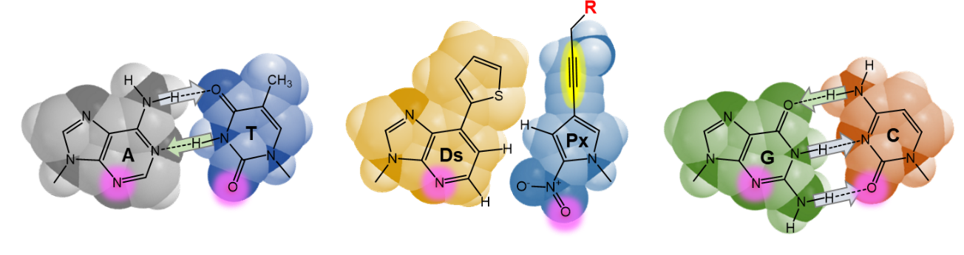
Six-Letter DNA Alphabet
We have developed an unnatural base pair system for six-letter PCR in ExSELEX. The hydrophobic Ds−Px pair can function as a third base pair with extremely high selectivity (more than 99.9%) in PCR. The Ds and Px bases significantly enhance physicochemical and structural diversities of DNA, facilitating the generation of XenoAptamers.
References
Unnatural Base Pair Development:
Curr. Opin. Chem. Biol., 2018., Angew. Chem. Int. Ed. Engl., 2017., ACS Synthetic Biol., 2016., Proc. Jpn. Acad. Ser. B Phys. Biol. Sci., 2012., Acc. Chem. Res., 2012., Nucleic Acids Res., 2012., Nucleic Acids Res., 2009., J. Am. Chem. Soc., 2007., Nature Methods, 2006., J. Am. Chem. Soc., 2005., J. Am. Chem. Soc., 2004., Nature Biotechnol., 2002., Proc. Natl. Acad. Sci. USA, 2001.
